Failed Sequencing Reaction
Identification
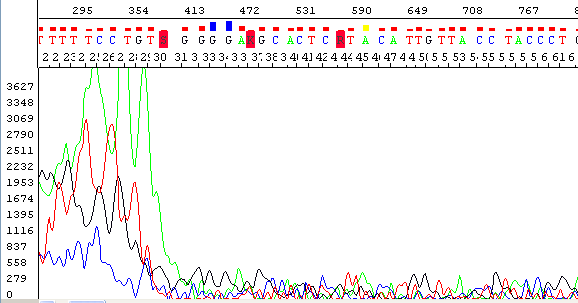
- Electropherogram has noisy or "messy" sequence peaks with low quality scores
- No or little signal in the raw data channels except for leftover dye at the beginning of the trace
- Signal strength in the raw channel usually below 100
- The sequence does not match either the expected sequence or other sequences in GenBank
Causes
- Insufficient/Poor quality DNA (particularly when using plasmids as the DNA template)
- Loss of the reaction during clean up (particularly when using ethanol precipitation as method of clean-up)
- Too much template DNA
- Absence of/mutation in primer binding site
- Poor water quality (may contain a sequencing inhibitor)
- Degraded or failed synthesis primer
- Degraded sequencing chemistry. Can occur if the BigDye chemistry is stored under the wrong conditions or is freeze-thawed too many times. Either the Taq DNA polymerase or dye labeled nucleotides can have degraded
Overcoming problem
Insufficient/Poor quality DNA
The best way to avoid this problem is to sequence a PCR amplified fragment of the plasmid insert instead of plasmid DNA itself. If this is not possible then it is recommended that a plasmid miniprep kit is used. It is also suggested that an ethanol precipitation is carried out after using the kit to further purify the DNA. Make sure the template is quantified accurately using a combination of agarose gel and spectrophotometer readings. Remember that we require 1ng/100bp of PCR product or 150-200ng of plasmid in the total 5µl submitted.
Loss of sequencing reaction during clean up
There are a number of kits available that can be used as a replacement for ethanol precipitation and we particularly recommend using Sephadex columns (see our Protocols section for information). One way of avoiding loss of the reaction DNA pellet when using the ethanol protocol is to add 1µl of a 20mg/ml solution of glycogen (Sigma G-1508) to the sequencing reaction before adding the ethanol. This helps make the pellet visible and the glycogen does not seem to interfere with the injection of the sequencing fragments onto the sequencer's capillaries.
Too much template DNA used
Make sure template is accurately quantified using a combination of agarose gel and spectrophotometer readings.
Primer binding issues
Check the sequence of the primer and template to make sure that the primer binding site is present. Make your own working stocks of primers rather than rely on other people to make them up correctly. It's vital that you add the correct amount of primer to your sample for processing. We require 3.2pmol primer in the total 5µl submitted (1µl of a 3.2µM stock).
Poor water quality
Inhibitors that can stop sequencing reactions can end up in lab water stocks. If you think this may be a problem then throw out the water and use a fresh stock.
Degraded primer
Don't use old diluted primer stocks. Store the primers in 10mM Tris/ 0.1 mM EDTA (pH 8.5) rather than water. If you have any doubt about the primer quality discard it and make up a fresh working solution.
Failed primer synthesis
If you suspect that the primer is poor quality check using PCR. Alternatively, if you have a control template that you know should work with the primer then this can be a good way of identifying primer problems.
Degraded sequencing chemistry
This can usually be prevented by storing the BigDye chemistry in small aliquots and avoiding repeated freeze/thaw cycles. If a batch of BigDye chemistry has not been used for some time, or there is any doubt about how it has been stored, then it is advisable to perform a control sequencing reaction before undertaking a large number of experimental reactions.


